Chapter 9 Performance Measures
Once we run a simulation, we end up with a pile of results to sort through. For example, Figure 9.1 depicts the distribution of average treatment effect estimates from the cluster-randomized experiment simulation, which we generated in Chapter 8. There are three different estimators, each with 1000 replications. Each histogram is an approximation of the sampling distribution of the estimator, meaning its distribution across repetitions of the data-generating process. With results such as these, the question before us is now how to evaluate how well each procedure works. If we are comparing several different estimators, we also need to determine which ones work better or worse than others. In this chapter, we look at a variety of performance measures that can answer these questions.
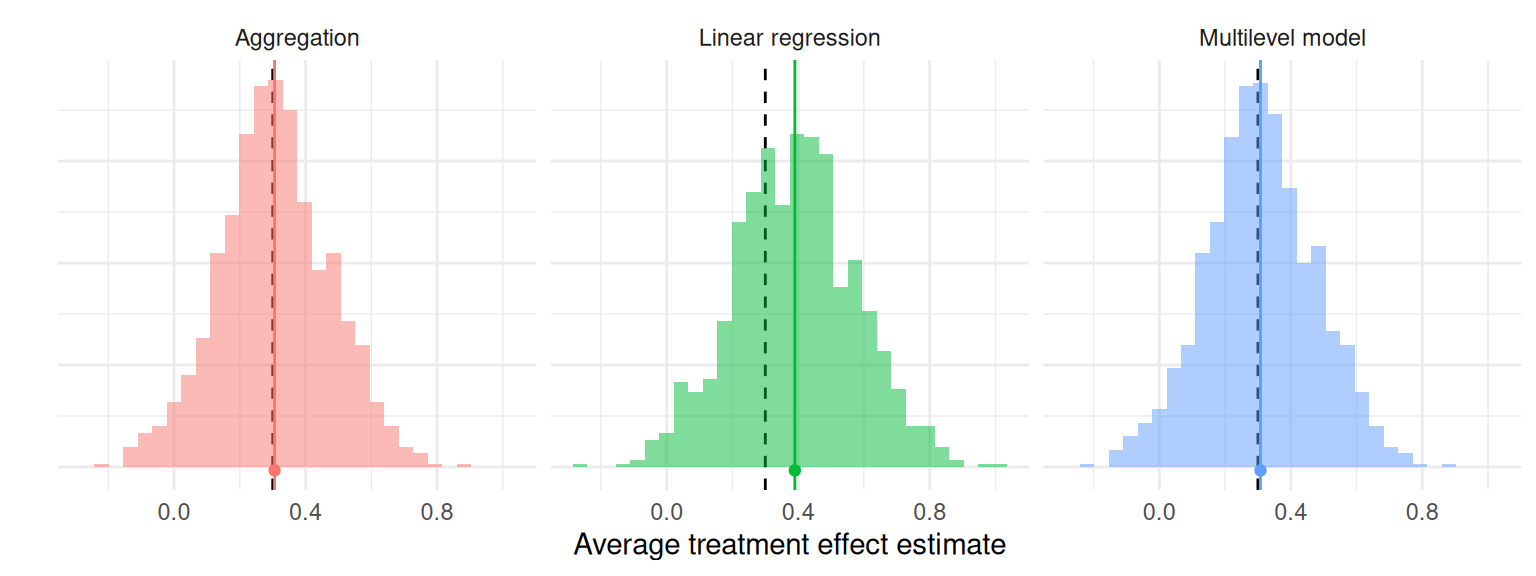
Figure 9.1: Sampling distribution of average treatment effect estimates from a cluster-randomized trial with a true average treatment effect of 0.3.
Performance measures are summaries of a sampling distribution that describe how an estimator or data analysis procedure behaves on average if we could repeat the data-generating process an infinite number of times. For example, the bias of an estimator is the difference between the average value of the estimator and the corresponding target parameter. Bias measures the central tendency of the sampling distribution, capturing how far off, on average, the estimator would be from the true parameter value if we repeated the data-generating process an infinite number of times. In Figure 9.1, black dashed lines mark the true average treatment effect of 0.3 and the colored vertical lines with circles at the end mark the means of the estimators. The distance between the colored lines and the black dashed lines corresponds to the bias of the estimator. This distance is nearly zero for the aggregation estimator and the multilevel model estimator, but larger for the linear regression estimator.
Different types of data-analysis results produce different types of information, and so the relevant set of performance measures depends on the type of data analysis result we are studying. For procedures that produce point estimates or point predictions, conventional performance measures include bias, variance, and root mean squared error. If the point estimates come with corresponding standard errors, then we may also want to evaluate how accurately the standard errors represent the true uncertainty of the point estimators; conventional performance measures for capturing this include the relative bias and relative root mean squared error of the variance estimator. For procedures that produce confidence intervals or other types of interval estimates, conventional performance measures include the coverage rate and average interval width. Finally, for inferential procedures that involve hypothesis tests (or more generally, classification tasks), conventional performance measures include Type I error rates and power. We describe each of these measures in Sections 9.1 through 9.4.
Performance measures are defined with respect to sampling distributions, or the results of applying a data analysis procedure to data generated according to a particular process across an infinite number of replications. In defining specific measures, we will follow statistical conventions to denote the mean, variance, and other moments of the sampling distribution. For a random variable \(T\), we will use the expectation operator \(\E(T)\) to denote the mean of the sampling distribution of \(T\), \(\M(T)\) to denote the median of its sampling distribution, \(\Var(T)\) to denote the variance of its sampling distribution, and \(\Prob()\) to denote probabilities of specific outcomes with respect to its sampling distribution. We will use \(\Q_p(T)\) to denote the \(p^{th}\) quantile of a distribution, which is the value \(x\) such that \(\Prob(T \leq x) = p\). With this notation, the median of a continuous distribution is equivalent to the 0.5 quantile: \(\M(T) = \Q_{0.5}(T)\).
For some simple combinations of data-generating processes and data analysis procedures, it may be possible to derive exact mathematical formulas for calculating some performance measures (such as exact mathematical expressions for the bias and variance of the linear regression estimator). But for many problems, the math is difficult or intractable—that’s why we do simulations in the first place. Simulations do not produce the exact sampling distribution or give us exact values of performance measures. Instead, simulations generate samples (usually large samples) from the the sampling distribution, and we can use these to compute estimates of the performance measures of interest. In Figure 9.1, we calculated the bias of each estimator by taking the mean of 1000 observations from its sampling distribution. If we were to repeat the whole set of calculations (with a different seed), then our bias results would shift slightly because they are imperfect estimates of the actual bias.
In working with simulation results, it is important to keep track of the degree of uncertainty in performance measure estimates. We call such uncertainty Monte Carlo error because it is the error arising from using a finite number of replications of the Monte Carlo simulation process. One way to quantify it is with the Monte Carlo standard error (MCSE), or the standard error of a performance estimate based on a finite number of replications. Just as when we analyze real data, we can apply statistical techniques to estimate the MCSE and even to generate confidence intervals for performance measures.
The magnitude of MCSE is driven by how many replications we use: if we only use a few, we will have noisy estimates of performance with large MCSEs; if we use millions of replications, the MCSE will usually be tiny. It is important to keep in mind that the MCSE is not measuring anything about how a data analysis procedure performs in general. It only describes how precisely we have approximated a performance criterion, an artifact of how we conducted the simulation. Moreover, MCSEs are under our control. Given a desired MCSE, we can determine how many replications we would need to ensure our performance estimates have the specified level of precision. Section 9.7 provides details about how to compute MCSEs for conventional performance measures, along with some discussion of general techniques for computing MCSE for less conventional measures.
9.1 Measures for Point Estimators
The most common performance measures used to assess a point estimator are bias, variance, and root mean squared error. Bias compares the mean of the sampling distribution to the target parameter. Positive bias implies that the estimator tends to systematically over-state the quantity of interest, while negative bias implies that it systematically under-shoots the quantity of interest. If bias is zero (or nearly zero), we say that the estimator is unbiased (or approximately unbiased). Variance (or its square root, the true standard error) describes the spread of the sampling distribution, or the extent to which it varies around its central tendency. All else equal, we would like estimators to have low variance (or to be more precise). Root mean squared error (RMSE) is a conventional measure of the overall accuracy of an estimator, or its average degree of error with respect to the target parameter. For absolute assessments of performance, an estimator with low bias, low variance, and thus low RMSE is desired. In making comparisons of several different estimators, one with lower RMSE is usually preferable to one with higher RMSE. If two estimators have comparable RMSE, then the estimator with lower bias would usually be preferable.
To define these quantities more precisely, let us consider a generic estimator \(T\) that is targeting a parameter \(\theta\). We call the target parameter the estimand. In most cases, in running our simulation we set the estimand \(\theta\) and then generate a (typically large) series of \(R\) datasets, for each of which \(\theta\) is the true target parameter. We then analyze each dataset, obtaining a sample of estimates \(T_1,...,T_R\). Formally, the bias, variance, and RMSE of \(T\) are defined as \[ \begin{aligned} \Bias(T) &= \E(T) - \theta, \\ \Var(T) &= \E\left[\left(T - \E (T)\right)^2 \right], \\ \RMSE(T) &= \sqrt{\E\left[\left(T - \theta\right)^2 \right]}. \end{aligned} \tag{9.1} \] These three measures are inter-connected. In particular, RMSE is the combination of (squared) bias and variance, as in \[ \left[\RMSE(T)\right]^2 = \left[\Bias(T)\right]^2 + \Var(T). \tag{9.2} \]
When conducting a simulation, we do not compute these performance measures directly but rather must estimate them using the replicates \(T_1,...,T_R\) generated from the sampling distribution. There is nothing very surprising about how we construct estimates of the performance measures. It is just a matter of substituting sample quantities in place of the expectations and variances. Specifically, we estimate bias by taking \[ \widehat{\Bias}(T) = \bar{T} - \theta, \tag{9.3} \] where \(\bar{T}\) is the arithmetic mean of the replicates, \(\bar{T} = \frac{1}{R}\sum_{r=1}^R T_r\). We estimate variance by taking the sample variance of the replicates, as \[ S_T^2 = \frac{1}{R - 1}\sum_{r=1}^R \left(T_r - \bar{T}\right)^2. \tag{9.4} \] \(S_T\) (the square root of \(S^2_T\)) is an estimate of the empirical standard error of \(T\), or the standard deviation of the estimator across an infinite set of replications of the data-generating process.11 We usually prefer to work with the empirical SE \(S_T\) rather than the sampling variance \(S_T^2\) because the former quantity has the same units as the target parameter. Finally, the RMSE estimate can be calculated as \[ \widehat{\RMSE}(T) = \sqrt{\frac{1}{R} \sum_{r = 1}^R \left( T_r - \theta\right)^2 }. \tag{9.5} \] Often, people talk about the MSE (Mean Squared Error), which is just the square of RMSE. Just as the true SE is usually easier to interpret than the sampling variance, units of RMSE are easier to interpret than the units of MSE.
It is important to recognize that the above performance measures depend on the scale of the parameter. For example, if our estimators are measuring a treatment impact in dollars, then the bias, SE, and RMSE of the estimators are all in dollars. (The variance and MSE would be in dollars squared, which is why we take their square roots to put them back on the more intepretable scale of dollars.)
In many simulations, the scale of the outcome is an arbitrary feature of the data-generating process, making the absolute magnitude of performance measures less meaningful. To ease interpretation of performance measures, it is useful to consider their magnitude relative to the baseline level of variation in the outcome. One way to achieve this is to generate data so the outcome has unit variance (i.e., we generate outcomes in standardized units). Doing so puts the bias, empirical standard error, and root mean squared error on the scale of standard deviation units, which can facilitate interpretation about what constitutes a meaningfully large bias or a meaningful difference in RMSE.
In addition to understanding the scale of these performance measures, it is also important to recognize that their magnitude depends on the metric of the parameter. A non-linear transformation of a parameter will generally lead to changes in the magnitude of the performance measures. For instance, suppose that \(\theta\) measures the proportion of time that something occurs. One natural way to transform this parameter would be to put it on the log-odds (logit) scale. However, because the log-odds transformation is non-linear, \[ \text{Bias}\left[\text{logit}(T)\right] \neq \text{logit}\left(\text{Bias}[T]\right), \qquad \text{RMSE}\left[\text{logit}(T)\right] \neq \text{logit}\left(\text{RMSE}[T]\right), \] and so on. This is a consequence of how these performance measures are defined. One might see this property as a limitation on the utility of using bias and RMSE to measure the performance of an estimator, because these measures can be quite sensitive to the metric of the parameter.
9.1.1 Comparing the Performance of the Cluster RCT Estimation Procedures
We now demonstrate the calculation of performance measures for the point estimators of average treatment effects in the cluster-RCT example. In Chapter 8, we generated a large set of replications of several different treatment effect estimators. Using these results, we can assess the bias, standard error, and RMSE of three different estimators of the ATE. These performance measures address the following questions:
- Is the estimator systematically off? (bias)
- Is it precise? (standard error)
- Does it predict well? (RMSE)
Let us see how the three estimators compare on these measures.
Are the estimators biased?
Bias is defined with respect to a target estimand. Here we assess whether our estimates are systematically different from the \(\gamma_1\) parameter, which we defined in standardized units by setting the standard deviation of the student-level distribution of the outcome equal to one. For these data, we generated data based on a school-level ATE parameter of 0.30 SDs.
ATE <- 0.30
runs %>%
group_by( method ) %>%
summarise(
mean_ATE_hat = mean( ATE_hat ),
bias = mean( ATE_hat ) - ATE
)## # A tibble: 3 × 3
## method mean_ATE_hat bias
## <chr> <dbl> <dbl>
## 1 Agg 0.300 -0.000166
## 2 LR 0.382 0.0824
## 3 MLM 0.312 0.0122There is no indication of major bias for aggregation or multi-level modeling. Linear regression, with a bias of about 0.09 SDs, appears about ten times as biased as the other estimators. This is because the linear regression is targeting the person-level average average treatment effect. The data-generating process of this simulation makes larger sites have larger effects, so the person-level average effect is going to be higher because those larger sites will count more. In contrast, our estimand is the school-level average treatment effect, or the simple average of each school’s true impact, which we have set to 0.30. The aggregation and multi-level modeling methods target this school-level average effect. If we had instead decided that the target estimand should be the person-level average effect, then we would find that linear regression is unbiased whereas aggregation and multi-level modeling are biased. This example illustrates how crucial it is to think carefully about the appropriate target parameter and to assess performance with respect to a well-justified and clearly articulated target.
Which method has the smallest standard error?
The empirical standard error measures the degree of variability in a point estimator. It reflects how stable our estimates are across replications of the data-generating process. We calculate the standard error by taking the standard deviation of the replications of each estimator. For purposes of interpretation, it is useful to compare the empirical standard errors to the variation in a benchmark estimator. Here, we treat the linear regression estimator as the benchmark and compute the magnitude of the empirical SEs of each method relative to the SE of the linear regression estimator:
true_SE <-
runs %>%
group_by( method ) %>%
summarise( SE = sd( ATE_hat ) ) %>%
mutate( per_SE = SE / SE[method=="LR"] )
true_SE## # A tibble: 3 × 3
## method SE per_SE
## <chr> <dbl> <dbl>
## 1 Agg 0.214 0.956
## 2 LR 0.224 1
## 3 MLM 0.213 0.953In a real data analysis, these standard errors are what we would be trying to approximate with a standard error estimator. Aggregation and multi-level modeling have SEs about 8% smaller than linear regression. For these data-generating conditions, aggregation and multi-level modeling are preferable to linear regression because they are more precise.
Which method has the smallest Root Mean Squared Error?
So far linear regression is not doing well: it has more bias and a larger standard error than the other two estimators. We can assess overall accuracy by combining these two quantities with the RMSE:
runs %>%
group_by( method ) %>%
summarise(
bias = mean( ATE_hat - ATE ),
SE = sd( ATE_hat ),
RMSE = sqrt( mean( (ATE_hat - ATE)^2 ) )
) %>%
mutate(
per_RMSE = RMSE / RMSE[method=="LR"]
)## # A tibble: 3 × 5
## method bias SE RMSE per_RMSE
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Agg -0.000166 0.214 0.214 0.897
## 2 LR 0.0824 0.224 0.238 1
## 3 MLM 0.0122 0.213 0.213 0.896We also include SE and bias for ease of reference.
RMSE takes into account both bias and variance. For aggregation and multi-level modeling, the RMSE is the same as the standard error, which makes sense because these estimators are not biased. For linear regression, the combination of bias plus increased variability yields a higher RMSE, with the standard error dominating the bias term (note how RMSE and SE are more similar than RMSE and bias). The difference between the estimators are pronounced because RMSE is the square root of the squared bias and squared standard error. Overall, aggregation and multi-level modeling have RMSEs around 17% smaller than linear regression—a consequential difference in accuracy.
9.1.2 Less Conventional Performance Measures
Depending on the model and estimation procedures being examined, a range of different measures might be used to assess estimator performance. For point estimation, we have introduced bias, variance and RMSE as three core measures of performance. However, all of these measures are sensitive to outliers in the sampling distribution. Consider an estimator that generally does well, except for an occasional large mistake. Because conventional measures are based on arithmetic averages, they will indicate that the estimator performs very poorly overall. Other measures such as the median bias and the median absolute deviation of \(T\) are less sensitive to outliers in the sampling distribution compared to the conventional measures. Estimating these measures will involve calculating sample quantiles of \(T_1,...,T_R\), which are functions of the sample ordered from smallest to largest. We will denote the \(r^{th}\) order statistic as \(T_{(r)}\) for \(r = 1,...,R\).
Median bias is an alternative measure of the central tendency of a sampling distribution. Positive median bias implies that more than 50% of the sampling distribution exceeds the quantity of interest, while negative median bias implies that more than 50% of the sampling distribution fall below the quantity of interest. Formally, \[ \text{Median-Bias}(T) = \M(T) - \theta \tag{9.6}. \] An estimator of median bias is computed using the sample median, as \[ \widehat{\text{Median-Bias}}(T) = M_T - \theta \tag{9.7} \] where \(M_T = T_{((R+1)/2)}\) if \(R\) is odd or \(M_T = \frac{1}{2}\left(T_{(R/2)} + T_{(R/2+1)}\right)\) if \(R\) is even.
Another robust measure of central tendency uses the \(p \times 100\%\)-trimmed mean, which ignores the estimates in the lowest and highest \(p\)-quantiles of the sampling distribution. Formally, the trimmed-mean bias is \[ \text{Trimmed-Bias}(T; p) = \E\left[ T \left| \Q_{p}(T) < T < \Q_{(1 - p)}(T) \right.\right] - \theta. \tag{9.8} \] Median bias is thus a special case of trimmed mean bias, with \(p = 0.5\). To estimate the trimmed bias, we take the mean of the middle \(1 - 2p\) fraction of the distribution \[ \widehat{\text{Trimmed-Bias}}(T; p) = \tilde{T}_{\{p\}} - \theta. \tag{9.9} \] where \[ \tilde{T}_{\{p\}} = \frac{1}{(1 - 2p)R} \sum_{r=pR + 1}^{(1-p)R} T_{(r)} \] For a symmetric sampling distribution, trimmed-mean bias will be the same as the conventional (mean) bias, but its estimator \(\tilde{T}_{\{p\}}\) will be less affected by outlying values (i.e., values of \(T\) very far from the center of the distribution) compared to \(\bar{T}\). However, if a sampling distribution is not symmetric, trimmed-mean bias become distinct performance measures, which put less emphasis on large errors compared to the conventional bias measure.
A further robust measure of central tendency is based on winsorizing the sampling distribution, or truncating all errors larger than a certain maximum size. Using a winsorized distribution amounts to arguing that you don’t care about errors beyond a certain size, so anything beyond a certain threshold will be treated the same as if it were exactly on the threshold. The threshold for truncation is usually defined relative to the first and third quartiles of the sampling distribution, along with a given span of the inter-quartile range. The thresholds for truncation are taken as \[ \begin{aligned} L_w &= \Q_{0.25}(T) - w \times (\Q_{0.75}(T) - \Q_{0.25}(T)) \\ U_w &= \Q_{0.75}(T) + w \times (\Q_{0.75}(T) - \Q_{0.25}(T)), \end{aligned} \] where \(\Q_{0.25}(T)\) and \(\Q_{0.75}(T)\) are the first and third quartiles of the distribution of \(T\), respectively, and \(w\) is the number of inter-quartile ranges below which an observation will be treated as an outlier.12 Let \(X = \min\{\max\{T, L_w\}, U_w\}\). The winsorized bias, variance, and RMSE are then defined using winsorized values in place of the raw values of \(T\), as \[ \begin{aligned} \text{Bias}(X) &= \E\left(X\right) - \theta \\ \Var\left(X\right) &= \E\left[\left(X - \E (T^{(w)})\right)^2 \right], \\ \RMSE\left(X\right) &= \sqrt{\E\left[\left(X - \theta\right)^2 \right]}. \end{aligned} \tag{9.10} \] To compute estimates of the winsorized performance criteria, we substitute sample quantiles \(T_{(R/4)}\) and \(T_{(3R/4)}\) in place of \(\Q_{0.25}(T)\) and \(\Q_{0.25}(T)\), respectively, to get estimated thresholds, \(\hat{L}_w\) and \(\hat{U}_w\), find \(\hat{X}_r = \min\{\max\{T_r, \hat{L}_w\}, \hat{U}_w\}\), and compute the sample performance measures using Equations (9.3), (9.4), and (9.5), but with \(\hat{X}\) in place of \(T_r\).
Alternative measures of the overall accuracy of an estimator can also be defined using quantiles.
For instance, an alternative to RMSE is to use the median absolute error (MAE), defined as
\[
\text{MAE} = \M\left(\left|T - \theta\right|\right).
\tag{9.11}
\]
Letting \(E_r = |T_r - \theta|\), the MAE can be estimated by taking the sample median of \(E_1,...,E_R\).
Many other robust measures of the spread of the sampling distribution are also available, including the Rosseeuw-Croux scale estimator \(Q_n\) [@Rousseeuw1993alternatives] and the biweight midvariance [@Wilcox2022introduction].
@Maronna2006robust provide a useful introduction to these measures and robust statistics more broadly.
The robustbase package [@robustbase] provides functions for calculating many of these robust statistics.
9.2 Measures for Variance Estimators
Statistics is concerned not only with how to estimate things, but also with understanding the extent of uncertainty in estimates of target parameters. These concerns apply in Monte Carlo simulation studies as well. In a simulation, we can simply compute an estimator’s actual properties. When we use an estimator with real data, we need to estimate its associated standard error and generate confidence intervals and other assessments of uncertainty. To understand if these uncertainty assessments work in practice, we need to evaluate not only the behavior of the estimator itself, but also the behavior of these associated quantities.
Commonly used measures for quantifying the performance of estimated standard errors include relative bias, relative standard error, and relative root mean squared error. These measures are defined in relative terms (rather than absolute ones) by comparing their magnitude to the true degree of uncertainty. Typically, performance measures are computed for variance estimators rather than standard error estimators. There are a few reasons for working with variance rather than standard error. First, in practice, so-called unbiased standard errors usually are not actually unbiased.13 For linear regression, for example, the classic standard error estimator is an unbiased variance estimator, but the standard error estimator is not exactly unbiased because \[ \E[ \sqrt{ V } ] \neq \sqrt{ \E[ V ] }. \] Variance is also the measure that gives us the bias-variance decomposition of Equation (9.2). Thus, if we are trying to determine whether MSE is due to instability or systematic bias, operating in this squared space may be preferable.
To make this concrete, let us consider a generic standard error estimator \(\widehat{SE}\) to go along with our generic estimator \(T\) of target parameter \(\theta\), and let \(V = \widehat{SE}^2\). We can simulate to obtain a large sample of standard errors, \(\widehat{SE}_1,...,\widehat{SE}_R\) and variance estimators \(V_r = \widehat{SE}_r^2\) for \(r = 1,...,R\). Formally, the relative bias, standard error, and RMSE of \(V\) are defined as \[ \begin{aligned} \text{Relative Bias}(V) &= \frac{\E(V)}{\Var(T)} \\ \text{Relative SE}(V) &= \frac{\Var(V)}{\Var(T)} \\ \text{Relative RMSE}(V) &= \frac{\sqrt{\E\left[\left(V - \Var(T)\right)^2 \right]}}{\Var(T)}. \end{aligned} \tag{9.12} \] In contrast to performance measures for \(T\), we define these measures in relative terms because the raw magnitude of \(V\) is not a stable or interpretable parameter. Rather, the sampling distribution of \(V\) will generally depend on many of the parameters of the data-generating process, including the sample size and any other design parameters. Defining bias in relative terms makes for a more interpretable metric: a value of 1 corresponds to exact unbiasedness of the variance estimator. Relative bias measures proportionate under- or over-estimation. For example, a relative bias of 1.12 would mean the standard error was, on average, 12% too large. We discuss relative performance measures further in Section 9.5.
To estimate these relative performance measures, we proceed by substituting sample quantities in place of the expectations and variances. In contrast to the performance measures for \(T\), we will not generally be able to compute the true degree of uncertainty exactly. Instead, we must estimate the target quantity \(\Var(T)\) using \(S_T^2\), the sample variance of \(T\) across replications. Denoting the arithmetic mean of the variance estimates as \[ \bar{V} = \frac{1}{R} \sum_{r=1}^R V_r \] and the sample variance as \[ S_V^2 = \frac{1}{R - 1}\sum_{r=1}^R \left(V_r - \bar{V}\right)^2, \] we estimate the relative bias, standard error, and RMSE of \(V\) using \[ \begin{aligned} \widehat{\text{Relative Bias}}(V) &= \frac{\bar{V}}{S_T^2} \\ \widehat{\text{Relative SE}}(V) &= \frac{S_V}{S_T^2} \\ \widehat{\text{Relative RMSE}}(V) &= \frac{\sqrt{\frac{1}{R}\sum_{r=1}^R\left(V_r - S_T^2\right)^2}}{S_T^2}. \end{aligned} \tag{9.13} \] These performance measures are informative about the properties of the uncertainty estimator \(V\) (or standard error \(\widehat{SE}\)), which have implications for the performance of other uncertainty assessments such as hypothesis tests and confidence intervals. Relative bias describes whether the central tendency of \(V\) aligns with the actual degree of uncertainty in the point estimator \(T\). Relative bias of less than 1 implies that \(V\) tends to under-state the amount of uncertainty, which will lead to confidence intervals that are overly narrow and do not cover the true parameter value at the desired rate. Relative bias greater than 1 implies that \(V\) tends to over-state the amount of uncertainty in the point estimator, making it seem like \(T\) is less precise than it truly is. Relative standard errors describe the variability of \(V\) in comparison to the true degree of uncertainty in \(T\)—the lower the better. A relative standard error of 0.5 would mean that the variance estimator has average error of 50% of the true uncertainty, implying that \(V\) will often be off by a factor of 2 compared to the true sampling variance of \(T\). Ideally, a variance estimator will have small relative bias, small relative standard errors, and thus small relative RMSE.
9.2.1 Satterthwaite degrees of freedom
Another more abstract measure of the stability of a variance estimator is its Satterthwaite degrees of freedom. For some simple statistical models such as classical analysis of variance and linear regression with homoskedastic errors, the variance estimator is computed by taking a sum of squares of normally distributed errors. In such cases, the sampling distribution of the variance estimator is a multiple of a \(\chi^2\) distribution, with degrees of freedom corresponding to the number of independent observations used to compute the sum of squares. In the context of analysis of variance problems, @Satterthwaite1946approximate described a method of approximating the variability of more complex statistics, involving linear combinations of sums of squares, by using a chi-squared distribution with a certain degrees of freedom. When applied to an arbitrary variance estimator \(V\), these degrees of freedom can be interpreted as the number of independent, normally distributed errors going into a sum of squares that would lead to a variance estimator that is equally precise as \(V\). More succinctly, these degrees of freedom correspond to the amount of independent observations used to estimate \(V\).
Following @Satterthwaite1946approximate, we define the degrees of freedom of \(V\) as \[ df = \frac{2 \left[\E(V)\right]^2}{\Var(V)}. \tag{9.14} \] We can estimate the degrees of freedom by taking \[ \widehat{df} = \frac{2 \bar{V}^2}{S_V^2}. \tag{9.15} \] For simple statistical methods in which \(V\) is based on a sum-of-squares of normally distributed errors, then the Satterthwaite degrees of freedom will be constant and correspond exactly to the number of independent observations in the sum of squares. Even with more complex methods, the degrees of freedom are interpretable: higher degrees of freedom imply that \(V\) is based on more observations, and thus will be a more precise estimate of the actual degree of uncertainty in \(T\).
9.2.2 Assessing SEs for the Cluster RCT Simulation
Returning to the cluster RCT example, we will assess whether our estimated SEs are about right by comparing the average estimated (squared) standard error versus the empirical sampling variance. Our standard errors are inflated if they are systematically larger than they should be, across the simulation runs. We will also look at how stable our variance estimates are by comparing their standard deviation to the empirical sampling variance and by computing the Satterthwaite degrees of freedom.
SE_performance <-
runs %>%
mutate( V = SE_hat^2 ) %>%
group_by( method ) %>%
summarise(
SE_sq = var( ATE_hat ),
V_bar = mean( V ),
rel_bias = V_bar / SE_sq,
S_V = sd( V ),
rel_SE_V = S_V / SE_sq,
df = 2 * mean( V )^2 / var( V )
)
SE_performance## # A tibble: 3 × 7
## method SE_sq V_bar rel_bias S_V rel_SE_V
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Agg 0.0457 0.0515 1.13 0.0175 0.384
## 2 LR 0.0500 0.0553 1.11 0.0231 0.461
## 3 MLM 0.0454 0.0510 1.12 0.0173 0.382
## # ℹ 1 more variable: df <dbl>The variance estimators for the aggregation estimator and multilevel model estimator appear to be a bit conservative on average, with relative bias of around 1.13, or about 13% higher than the true sampling variance.
The column labelled rel_SE_V reports how variable the variance estimators are relative to the true sampling variances of the estimators.
The column labelled df reports the Satterthwaite degrees of freedom of each variance estimator.
Both of these measures indicate that the linear regression variance estimator is less stable than the other methods, with around 6 fewer degrees of freedom.
The linear regression method uses a cluster-robust variance estimator, which is known to be a bit unstable [@cameronPractitionerGuideClusterRobust2015].
Overall, it is a bad day for linear regression.
9.3 Measures for Confidence Intervals
Some estimation procedures provide confidence intervals (or confidence sets) which are ranges of values, or interval estimators, that should include the true parameter value with a specified confidence level. For a 95% confidence level, the interval should include the true parameter in 95% replications of the data-generating process. However, with the exception of some simple methods and models, methods for constructing confidence intervals usually involve approximations and simplifying assumptions, so their actual coverage rate might deviate from the intended confidence level.
We typically measure confidence interval performance along two dimensions: coverage rate and expected width. Suppose that the confidence interval is for the target parameter \(\theta\) and has intended coverage level \(\beta\) for \(0 < \beta < 1\). Denote the lower and upper end-points of the \(\beta\)-level confidence interval as \(A\) and \(B\). \(A\) and \(B\) are random quantities—they will differ each time we compute the interval on a different replication of the data-generating process. The coverage rate of a \(\beta\)-level interval estimator is the probability that it covers the true parameter, formally defined as \[ \text{Coverage}(A,B) = \Prob(A \leq \theta \leq B). \tag{9.16} \] For a well-performing interval estimator, \(\text{Coverage}\) will at least \(\beta\) and, ideally will not exceed \(\beta\) by too much. The expected width of a \(\beta\)-level interval estimator is the average difference between the upper and lower endpoints, formally defined as \[ \text{Width}(A,B) = \E(B - A). \tag{9.17} \] Smaller expected width means that the interval tends to be narrower, on average, and thus more informative about the value of the target parameter.
In practice, we approximate the coverage and width of a confidence interval by summarizing across replications of the data-generating process. Let \(A_r\) and \(B_r\) denote the lower and upper end-points of the confidence interval from simulation replication \(r\), and let \(W_r = B_r - A_r\), all for \(r = 1,...,R\). The coverage rate and expected length measures can be estimated as \[ \begin{aligned} \widehat{\text{Coverage}}(A,B) &= \frac{1}{R}\sum_{r=1}^R I(A_r \leq \theta \leq B_r) \\ \widehat{\text{Width}}(A,B) &= \frac{1}{R} \sum_{r=1}^R W_r = \frac{1}{R} \sum_{r=1}^R \left(B_r - A_r\right). \end{aligned} \tag{9.18} \] Following a strict statistical interpretation, a confidence interval performs acceptably if it has actual coverage rate greater than or equal to \(\beta\). If multiple methods satisfy this criterion, then the method with the lowest expected width would be preferable. Some analysts prefer to look at lower and upper coverage separately, where lower coverage is \(\Prob(A \leq \theta)\) and upper coverage is \(\Prob(\theta \leq B)\).
In many instances, confidence intervals are constructed using point estimators and uncertainty estimators. For example, a conventional Wald-type confidence interval is centered on a point estimator, with end-points taken to be a multiple of an estimated standard error below and above the point estimator: \[ A = T - c \times \widehat{SE}, \quad B = T + c \times \widehat{SE} \] for some critical value \(c\) (e.g.,for a normal critical value with a \(\beta = 0.95\) confidence level, \(c = 1.96\)). Because of these connections, confidence interval coverage will often be closely related to the performance of the point estimator and variance estimator. Biased point estimators will tend to have confidence intervals with coverage below the desired level because they are not centered in the right place. Likewise, variance estimators that have relative bias below 1 will tend to produce confidence intervals that are too short, leading to coverage below the desired level. Thus, confidence interval coverage captures multiple aspects of the performance of an estimation procedure.
9.3.1 Confidence Intervals in the Cluster RCT Simulation
Returning to the CRT simulation, we will examine the coverage and expected width of normal Wald-type confidence intervals for each of the estimators under consideration.
To do this, we first have to calculate the confidence intervals because we did not do so in the estimation function.
We compute a normal critical value for a \(\beta = 0.95\) confidence level using qnorm(0.975), then compute the lower and upper end-points using the point estimators and estimated standard errors:
runs_CIs <-
runs %>%
mutate(
A = ATE_hat - qnorm(0.975) * SE_hat,
B = ATE_hat + qnorm(0.975) * SE_hat
)Now we can estimate the coverage rate and expected width of these confidence intervals:
runs_CIs %>%
group_by( method ) %>%
summarise(
coverage = mean( A <= ATE & ATE <= B ),
width = mean( B - A )
)## # A tibble: 3 × 3
## method coverage width
## <chr> <dbl> <dbl>
## 1 Agg 0.948 0.877
## 2 LR 0.92 0.903
## 3 MLM 0.945 0.872The coverage rate is close to the desired level of 0.95 for the multilevel model and aggregation estimators, but it is around 5 percentage points too low for linear regression. The lower-than-nominal coverage level occurs because of the bias of the linear regression point estimator. The linear regression confidence intervals are also a bit wider than the other methods due to the larger sampling variance of its point estimator and higher variability (lower degrees of freedom) of its standard error estimator.
The normal Wald-type confidence intervals we have examined here are based on fairly rough approximations. In practice, we might want to examine more carefully constructed intervals such as ones that use critical values based on \(t\) distributions or ones constructed by profile likelihood. Especially in scenarios with a small or moderate number of clusters, such methods might provide better intervals, with coverage closer to the desired confidence level. See Exercise 9.10.4.
9.4 Measures for Inferential Procedures (Hypothesis Tests)
Hypothesis testing entails first specifying a null hypothesis, such as that there is no difference in average outcomes between two experimental groups. One then collects data and evaluates whether the observed data is compatible with the null hypothesis. Hypothesis test results are often describes in terms of a \(p\)-value, which measures how extreme or surprising a feature of the observed data (a test statistic) is relative to what one would expect if the null hypothesis is true. A small \(p\)-value (such as \(p < .05\) or \(p < .01\)) indicates that the observed data would be unlikely to occur if the null is true, leading the researcher to reject the null hypothesis. Alternately, testing procedures might be formulated by comparing a test statistic to a specified critical value; a test statistic exceeding the critical value would lead the researcher to reject the null.
Hypothesis testing procedures aim to control the level of false positives, corresponding to the probability that the null hypothesis is rejected when it holds in truth. The level of a testing procedure is often denoted as \(\alpha\), and it has become conventional in many fields to conduct tests with a level of \(\alpha = .05\).14 Just as in the case of confidence intervals, hypothesis testing procedures can sometimes be developed that will have false positive rates exactly equal to the intended level \(\alpha\). However, in many other problems, hypothesis testing procedures involve approximations or assumption violations, so that the actual rate of false positives might deviate from the intended \(\alpha\)-level. When we evaluate a hypothesis testing procedure, we are concerned with two primary measures of performance: validity and power.
9.4.1 Validity
Validity pertains to whether we erroneously reject a true null more than we should. An \(\alpha\)-level testing procedure is valid if it has no more than an \(\alpha\) chance of rejecting the null, when the null is true. If we were using the conventional \(\alpha = .05\) level, then a valid testing procedure will reject the null in only 50 of 1000 replications of a data-generating process where the null hypothesis actually holds true.
To assess validity, we will need to specify a data generating process where the null hypothesis holds (e.g., where there is no difference in average outcomes between experimental groups). We then generate a large series of data sets with a true null, conduct the testing procedure on each dataset and record the \(p\)-value or critical value, then score whether we reject the null hypothesis. In practice, we may be interested in evaluating a testing procedure by exploring data generation processes where the null is true but other aspects of the data (such as outliers, skewed outcome distributions, or small sample size) make estimation difficult, or where auxiliary assumptions of the testing procedure are violated. Examining such data-generating processes allows us to understand if our methods are robust to patterns that might be encountered in real data analysis. The key to evaluating the validity of a procedure is that, for whatever data-generating process we examine, the null hypothesis must be true.
9.4.2 Power
Power is concerned with the chance that we notice when an effect or a difference exists—that is, the probability of rejecting the null hypothesis when it does not actually hold. Compared to validity, power is a more nuanced concept because larger effects will clearly be easier to notice than smaller ones, and more blatant violations of a null hypothesis will be easier to identify than subtle ones. Furthermore, the rate at which we can detect violations of a null will depend on the \(\alpha\) level of the testing procedure. A lower \(\alpha\) level will make for a less sensitive test, requiring stronger evidence to rule out a null hypothesis. Conversely, a higher \(\alpha\) level will reject more readily, leading to higher power but at a cost of increased false positives.
In order to evaluate the power of a testing procedure by simulation, we will need to generate data where there is something to detect. In other words, we will need to ensure that the null hypothesis is violated (and that some specific alternative hypothesis of interest holds). The process of evaluating the power of a testing procedure is otherwise identical to that for evaluating its validity: generate many datasets, carry out the testing procedure, and track the rate at which the null hypothesis is rejected. The only difference is the conditions under which the data are generated.
We find it useful to think of power as a function rather than as a single quantity because its absolute magnitude will generally depend on the sample size of a dataset and the magnitude of the effect of interest. Because of this, power evaluations will typically involve examining a sequence of data-generating scenarios with varying sample size or varying effect size. Further, if our goal is to evaluate several different testing procedures, the absolute power of a procedure will be of less concern than the relative performance of one procedure compared to another.
9.4.3 Rejection Rates
When evaluating either validity or power, the main performance measure is the rejection rate of the hypothesis test. Letting \(P\) be the p-value from a procedure for testing the null hypothesis that a parameter \(\theta = 0\), generated under a data-generating process with parameter \(\theta\) (which could in truth be zero or non-zero). The rejection rate is then \[ \rho_\alpha(\theta) = \Prob(P < \alpha) \tag{9.19} \] When data are simulated from a process in which the null hypothesis is true, then the rejection rate is equivalent to the Type-I error rate of the test, which should ideally be near the desired \(\alpha\) level. When the data are simulated from a process in which the null hypothesis is violated, then the rejection rate is equivalent to the power of the test (for the given alternate hypothesis specified in the data-generating process). Ideally, a testing procedure should have actual Type-I error equal to the nominal level \(\alpha\) (this is the definition of validity), but such exact tests are rare.
To estimate the rejection rate of a test, we calculate the proportion of replications where the test rejects the null hypothesis. Letting \(P_1,...,P_R\) be the p-values simulated from \(R\) replications of a data-generating process with true parameter \(\theta\), we estimate the rejection rate by calculating \[ r_\alpha(\theta) = \frac{1}{R} \sum_{r=1}^R I(P_r < \alpha). \tag{9.20} \] It may be of interest to evaluate the performance of the test at several different \(\alpha\) levels. For instance, @brown1974SmallSampleBehavior evaluated the Type-I error rates and power of their tests using \(\alpha = .01\), \(.05\), and \(.10\). Simulating the \(p\)-value of the test makes it easy to estimate rejection rates for multiple \(\alpha\) levels, since we simply need to apply Equation (9.20) for several values of \(\alpha\). When simulating from a data-generating process where the null hypothesis holds, one can also plot the empirical cumulative distribution function of the \(p\)-values; for an exactly valid test, the \(p\)-values should follow a standard uniform distribution with a cumulative distribution falling along the \(45^\circ\) line.
Methodologists hold a variety of perspectives on how close the actual Type-I error rate should be in order to qualify as suitable for use in practice. Following a strict statistical definition, a hypothesis testing procedure is said to be level-\(\alpha\) if its actual Type-I error rate is always less than or equal to \(\alpha\), for any specific conditions of a data-generating process. Among a collection of level-\(\alpha\) testing procedures, we would prefer the one with highest power. If looking only at null rejection rates, then the test with Type-I error closest to \(\alpha\) would usually be preferred. However, some scholars prefer to use a less stringent criterion, where the Type-I error rate of a testing procedure would be considered acceptable if it is within 50% of the desired \(\alpha\) level. For instance, a testing procedure with \(\alpha = .05\) would be considered acceptable if its Type-I error is no more than 7.5%; with \(\alpha = .01\), it would be considered acceptable if its Type-I error is no more than 1.5%.
9.4.4 Inference in the Cluster RCT Simulation
Returning to the cluster RCT simulation, we will evaluate the validity and power of hypothesis tests for the average treatment effect based on each of the three estimation methods. The data used in previous sections of the chapter was simulated under a process with a non-null treatment effect parameter (equal to 0.3 SDs), so the null hypothesis of zero average treatment effect does not hold. Thus, the rejection rates for this scenario correspond to estimates of power. We compute the rejection rate for tests with an \(\alpha\) level of \(.05\):
## # A tibble: 3 × 2
## method power
## <chr> <dbl>
## 1 Agg 0.241
## 2 LR 0.32
## 3 MLM 0.263For this particular scenario, none of the tests have especially high power, and the linear regression estimator apparently has higher power than the aggregation method and the multi-level model.
To make sense of this power pattern, we need to also consider the validity of the testing procedures.
We can do so by re-running the simulation using code we constructed in Chapter 8 using the simhelpers package.
To evaluate the Type-I error rate of the tests, we will set the average treatment effect parameter to zero by specifying ATE = 0:
set.seed( 404044 )
runs_val <- sim_cluster_RCT(
reps = 1000,
J = 20, n_bar = 30, alpha = 0.75,
gamma_1 = 0, gamma_2 = 0.5,
sigma2_u = 0.2, sigma2_e = 0.8
)Assessing validity involves repeating the exact same rejection rate calculations as we did for power:
## # A tibble: 3 × 2
## estimator power
## <chr> <dbl>
## 1 MLM 0.03
## 2 OLS 0.054
## 3 agg 0.029The Type-I error rates of the tests for the aggregation and multi-level modeling approaches are around 5%, as desired. The test for the linear regression estimator has Type-I error above the specified \(\alpha\)-level due to the upward bias of the point estimator used in constructing the test. The elevated rejection rate might be part of the reason that the linear regression test has higher power than the other procedures. It is not entirely fair to compare the power of these testing procedures, because one of them has Type-I error in excess of the desired level.
As discussed above, linear regression targets the person-level average treatment effect. In the scenario we simulated for evaluating validity, the person-level average effect is not zero because we have specified a non-zero impact heterogeneity parameter (\(\gamma_2=0.2\)), meaning that the school-specific treatment effects vary around 0. To see if this is why the linear regression test has an inflated Type-I error rate, we could re-run the simulation using settings where both the school-level and person-level average effects are truly zero.
9.5 Relative or Absolute Measures?
In considering performance measures for point estimators, we have defined the measures in terms of differences (bias, median bias) and average deviations (variance and RMSE), all of which are on the scale of the target parameter. In contrast, for evaluating estimated standard errors we have defined measures in relative terms, calculated as ratios of the target quantity rather than as differences. In the latter case, relative measures are justified because the target quantity (the true degree of uncertainty) is always positive and is usually strongly affected by design parameters of the data-generating process. Is it ever reasonable to use relative measures for point estimators? If so, how should we decide whether to use relative or absolute measures?
Many published simulation studies have used relative performance measures for evaluating point estimators. For instance, studies might use relative bias or relative RMSE, defined as \[ \begin{aligned} \text{Relative }\Bias(T) &= \frac{\E(T)}{\theta}, \\ \text{Relative }\RMSE(T) &= \frac{\sqrt{\E\left[\left(T - \theta\right)^2 \right]}}{\theta}. \end{aligned} \tag{9.21} \] and estimated as \[ \begin{aligned} \widehat{\text{Relative }\Bias(T)} &= \frac{\bar{T}}{\theta}, \\ \widehat{\text{Relative }\RMSE(T)} &= \frac{\widehat{RMSE}(T)}{\theta}. \end{aligned} \tag{9.22} \] As justification for evaluating bias in relative terms, authors often appeal to @hoogland1998RobustnessStudiesCovariance, who suggested that relative bias of under 5% (i.e., relative bias falling between 0.95 and 1.05) could be considered acceptable for an estimation procedure. However, @hoogland1998RobustnessStudiesCovariance were writing about a very specific context—robustness studies of structural equation modeling techniques—that have parameters of a particular form. In our view, their proposed rule-of-thumb is often generalized far beyond the circumstances where it might be defensible, including to problems where it is clearly arbitrary and inappropriate.
A more principled approach to choosing between absolute and relative measures is to consider how the magnitude of the measure changes across different values of the target parameter \(\theta\). If the estimand of interest is a location parameter, then shifting \(\theta\) by 0.1 or by 10.1 would not usually lead to changes in the magnitude of bias, variance, or RMSE. The relationship between bias and the target parameter might be similar to Scenario A in Figure 9.2, where bias is roughly constant across a range of different values of \(\theta\). Focusing on relative measures in this scenario would lead to a much more complicated story because different values of \(\theta\) will produce drastically different values, ranging from nearly unbiased to nearly infinite bias (for \(\theta\) very close to zero).
Another possibility is that shifting \(\theta\) by 0.1 or 10.1 will lead to proportionate changes in the magnitude of bias, variance, or RMSE. The relationship between bias and the target parameter might be similar to Scenario B in 9.2, where bias is is roughly a constant multiple of the target parameter \(\theta\). Focusing on relative measures in this scenario is useful because it leads to a simple story: relative bias is always around 1.12 across all values of \(\theta\), even though the raw bias varies considerably. We would usually expect this type of pattern to occur for scale parameters.
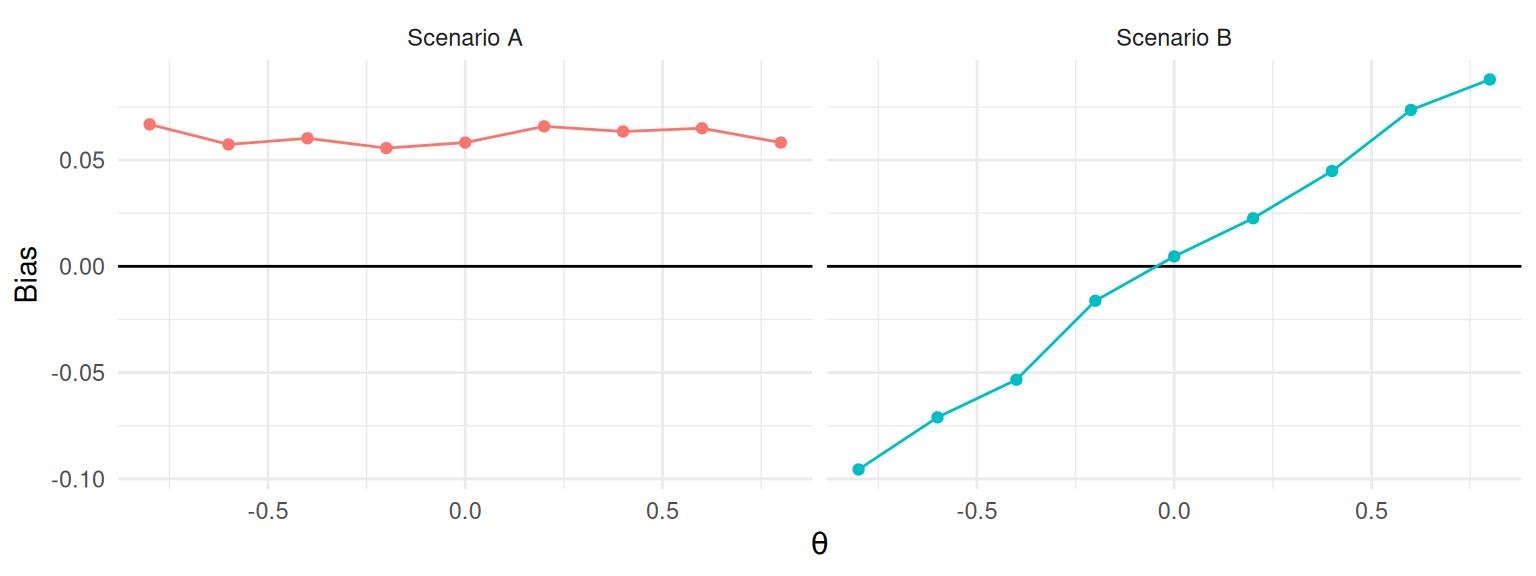
Figure 9.2: Hypothetical relationships between bias and a target parameter \(\theta\). In Scenario A, bias is unrelated to \(\theta\) and absolute bias is a more appropriate measure. In Scenario B, bias is proportional to \(\theta\) and relative bias is a more appropriate measure.
How do we know which of these scenarios is a better match for a particular problem? For some estimators and data-generating processes, it may be possible to analyze a problem with statistical theory and examine the how bias or variance would be expected to change as a function of \(\theta\). However, many problems are too complex to be tractable. Another, much more feasible route is to evaluate performance for multiple values of the target parameter. As done in many simulation studies, we can simulate sampling distributions and calculate performance measures (in raw terms) for several different values of a parameter, selected so that we can distinguish between constant and multiplicative relationships. Then, in analyzing the simulation results, we can generate graphs such as those in Figure 9.2 to understand how performance changes as a function of the target parameter. If the absolute bias is roughly the same for all values of \(\theta\) (as in Scenario A), then it makes sense to report absolute bias as the summary performance criterion. On the other hand, if the bias grows roughly in proportion to \(\theta\) (as in Scenario B), then relative bias might be a better summary criterion.
9.5.1 Performance relative to a benchmark estimator
Another way to define performance measures in relative terms to by taking the ratio of the performance measure for one estimator over the performance measure for a benchmark estimator. We have already demonstrated this approach in calculating performance measures for the cluster RCT example (Section 9.1.1), where we used the linear regression estimator as the benchmark against which to compare the other estimators. This approach is natural in simulations that involve comparing the performance of multiple estimators and where one of the estimators could be considered the current standard or conventional method.
Comparing the performance of one estimator relative to another can be especially useful when examining measures whose magnitude varies drastically across design parameters. For most statistical methods, we would usually expect precision and accuracy to improve (variance and RMSE to decrease) as sample size increases. Comparing estimators in terms of relative precision or relative accuracy may make it easier to identify consistent patterns in the simulation results. For instance, this approach might allow us to summarize findings by saying that “the aggregation estimator has standard errors that are consistently 6-10% smaller than the standard errors of the linear regression estimator.” This is much easier to interpret than saying that “aggregation has standard errors that are around 0.01 smaller than linear regression, on average.” In the latter case, it is very difficult to determine whether a difference of 0.01 is large or small, and focusing on an average difference conceals relevant variation across scenarios involving different sample sizes.
Comparing performance relative to a benchmark method can be an effective tool, but it also has potential drawbacks. Because these relative performance measures are inherently comparative, higher or lower ratios could either be due to the behavior of the method of interest (the numerator) or due to the behavior of the benchmark method (the denominator). Ratio comparisons are also less effective for performance measures that are on a constrained scale, such as power. If we have a power of 0.05, and we improve it to 0.10, we have doubled our power, but if it is 0.10 and we increase to 0.15, we have only increased by 50%. Ratios can also be very deceiving when the denominator quantity is near zero or when it can take on either negative or positive values; this can be a problem when examining bias relative to a benchmark estimator. Because of these drawbacks, it is prudent to compute and examine performance measures in absolute terms in addition to examining relative comparisons between methods.
9.6 Estimands Not Represented By a Parameter
In our Cluster RCT example, we focused on the estimand of the school-level ATE, represented by the model parameter \(\gamma_1\). What if we were instead interested in the person-level average effect? This estimand does not correspond to any input parameter in our data generating process. Instead, it is defined implicitly by a combination of other parameters. In order to compute performance characteristics such as bias and RMSE, we would need to calculate the parameter based on the inputs of the data-generating processes. There are at least three possible ways to accomplish this.
One way is to use mathematical distribution theory to compute an implied parameter.
Our target parameter will be some function of the parameters and random variables in the data-generating process, and it may be possible to evaluate that function algebraically or numerically (i.e., using numerical integration functions such as integrate()).
This can be a very worthwhile exercise if it provides insights into the relationship between the target parameter and the inputs of the data-generating process.
However, this approach requires knowledge of distribution theory, and it can get quite complicated and technical.15
Other approaches are often feasible and more closely aligned with the tools and techniques of Monte Carlo simulation.
An alternative approach is to simply generate a massive dataset—so large that it can stand in for the entire data-generating model—and then simply calculate the target parameter of interest in this massive dataset. In the cluster-RCT example, we can apply this strategy by generating data from a very large number of clusters and then simply calculating the true person-average effect across all generated clusters. If the dataset is big enough, then the uncertainty in this estimate will be negligible compared to the uncertainty in our simulation.
We implement this approach as follows, generating a dataset with 100,000 clusters:
dat <- gen_cluster_RCT(
n_bar = 30, J = 100000,
gamma_1 = 0.3, gamma_2 = 0.5,
sigma2_u = 0.20, sigma2_e = 0.80,
alpha = 0.75
)
ATE_person <- mean( dat$Yobs[dat$Z==1] ) - mean( dat$Yobs[dat$Z==0] )
ATE_person## [1] 0.3919907The extremely precise estimate of the person-average effect is 0.39, which is consistent with what we would expect given the bias we saw earlier for the linear model.
If we recalculate performance measures for all of our estimators with respect to the ATE_person estimand, the bias and RMSE of our estimators will shift but the standard errors will stay the same as in previous performance calculations using the school-level average effect:
performance_person_ATE <-
runs %>%
group_by( method ) %>%
summarise(
bias = mean( ATE_hat ) - ATE_person,
SE = sd( ATE_hat ),
RMSE = sqrt( mean( (ATE_hat - ATE_person)^2 ) )
) %>%
mutate( per_RMSE = RMSE / RMSE[method=="LR"] )
performance_person_ATE## # A tibble: 3 × 5
## method bias SE RMSE per_RMSE
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Agg -0.0922 0.214 0.233 1.04
## 2 LR -0.00963 0.224 0.224 1
## 3 MLM -0.0798 0.213 0.227 1.02For the person-weighted estimand, the aggregation estimator and multilevel model are biased but the linear regression estimator is unbiased. However, the aggregation estimator and multilevel model estimator still have smaller standard errors than the linear regression estimator. RMSE now captures the trade-off between bias and reduced variance. Overall, aggregation and multilevel modeling have RMSE that is around 3% larger than linear regression.
A further approach for calculating ATE_person would be to record the true person average effect of the dataset with each simulation iteration, and then average the sample-specific parameters at the end.
The overall average of the dataset-specific ATE_person parameters corresponds to the population person-level ATE.
This approach is equivalent to generating a single massive dataset—we just generate it piece by piece.
To implement this approach, we would need to modify the data-generating function gen_cluster_RCT() to track the additional information.
For instance, we might calculate
and then include tx_effect along with Yobs and Z as a column in our dataset.
This approach is quite similar to directly calculating potential outcomes, as discussed in Chapter 21.
After modifying the data-generating function, we will also need to modify the analysis function(s) to record the sample-specific treatment effect parameter. We might have, for example:
analyze_data = function( dat ) {
MLM <- analysis_MLM( dat )
LR <- analysis_OLS( dat )
Agg <- analysis_agg( dat )
res <- bind_rows(
MLM = MLM, LR = LR, Agg = Agg,
.id = "method"
)
res$ATE_person <- mean( dat$tx_effect )
return( res )
}Now when we run our simulation, we will have a column corresponding to the true person-level average treatment effect for each dataset. We could then take the average of these value across replications to estimate the true person average treatment effect in the population, and then use this as the target parameter for performance calculations.
An estimand not represented by any single input parameter is more difficult to work with than one that corresponds directly to an input parameter. Still, it is feasible to examine such estimands with a bit of forethought and careful programming. The key is to be clear about what you are trying to estimate because the performance of an estimator depends critically on the estimand against which it is compared.
9.7 Uncertainty in Performance Estimates (the Monte Carlo Standard Error)
The performance measures we have described are all defined with respect to the sampling distribution of an estimator, or its distribution across an infinite number of replications of the data-generating process. Of course, simulations will only involve a finite set of replications, based on which we calculate estimates of the performance measures. These estimates involve some Monte Carlo error because they are based on a limited number of replications. It is important to understand the extent of Monte Carlo error when interpreting simulation results, so we need methods for asssessing this source of uncertainty.
To account for Monte Carlo error, we can think of our simulation results as a sample from a population. Each replication is an independent and identically distributed draw from the population of the sampling distribution. Once we frame the problem in these terms, standard statistical techniques for independent and identically distributed random variables can be applied to calculate standard errors. We call these standard errors Monte Carlo Simulation Errors, or MCSEs. For most of the performance measures, closed-form expressions are available for calculating MCSEs. For a few of the measures, we can apply techniques such as the jackknife to calculate reasonable approximations for MCSEs.
9.7.1 Conventional measures for point estimators
For the measures that we have described for evaluating point estimators, Monte Carlo standard errors can be calculated using conventional formulas.16 Recall that we have a point estimator \(T\) of a target parameter \(\theta\), and we calculate the mean of the estimator \(\bar{T}\) and its sample standard deviation \(S_T\) across \(R\) replications of the simulation process. In addition, we will need to calculate the standardized skewness and kurtosis of \(T\) as
\[ \begin{aligned} \text{Skewness (standardized):} & &g_T &= \frac{1}{R S_T^3}\sum_{r=1}^R \left(T_r - \bar{T}\right)^3 \\ \text{Kurtosis (standardized):} & &k_T &= \frac{1}{R S_T^4} \sum_{r=1}^R \left(T_r - \bar{T}\right)^4. \end{aligned} \tag{9.23} \]
The bias of \(T\) is estimated as \(\bar{T} - \theta\), so the MCSE for bias is equal the MCSE of \(\bar{T}\). It can be estimated as \[ MCSE\left(\widehat{\Bias}(T)\right) = \sqrt{\frac{S_T^2}{R}}. \tag{9.24} \] The sampling variance of \(T\) is estimated as \(S_T^2\), with MCSE of \[ MCSE\left(\widehat{\Var}(T)\right) = S_T^2 \sqrt{\frac{k_T - 1}{R}}. \tag{9.25} \] The empirical standard error (the square root of the sampling variance) is estimated as \(S_T\). Using a delta method approximation17, the MCSE of \(S_T\) is \[ MCSE\left(S_T\right) = \frac{S_T}{2}\sqrt{\frac{k_T - 1}{R}}. \tag{9.26} \]
We estimate RMSE using Equation (9.5), which can also be written as \[ \widehat{\RMSE}(T) = \sqrt{(\bar{T} - \theta)^2 + \frac{R - 1}{R} S_T^2}. \] An MCSE for the estimated mean squared error (the square of RMSE) is \[ MCSE( \widehat{MSE} ) = \sqrt{\frac{1}{R}\left[S_T^4 (k_T - 1) + 4 S_T^3 g_T\left(\bar{T} - \theta\right) + 4 S_T^2 \left(\bar{T} - \theta\right)^2\right]}. \tag{9.27} \] Again following a delta method approximation, a MCSE for the RMSE is \[ MCSE( \widehat{RMSE} ) = \frac{\sqrt{\frac{1}{R}\left[S_T^4 (k_T - 1) + 4 S_T^3 g_T\left(\bar{T} - \theta\right) + 4 S_T^2 \left(\bar{T} - \theta\right)^2\right]}}{2 \times \widehat{RMSE}}. \tag{9.28} \]
Section 9.5 discussed circumstances where we might prefer to calculate performance measures in relative rather than absolute terms. For measures that are calculated by dividing a raw measure by the target parameter, the MCSE for the relative measure is simply the MCSE for the raw measure divided by the target parameter. For instance, the MCSE of relative bias \(\bar{T} / \theta\) is \[ MCSE\left( \frac{\bar{T}}{\theta} \right) = \frac{1}{\theta} MCSE(\bar{T}) = \frac{S_T}{\theta \sqrt{R}}. \tag{9.29} \] MCSEs for relative variance and relative RMSE follow similarly.
9.7.2 Less conventional measures for point estimators
In Section 9.1.2 we described several alternative performance measures for evaluating point estimators, which are less commonly used but are more robust to outliers compared to measures such as bias and variance. MCSEs for these less conventional measures can be obtained using results from the theory of robust statistics [@Hettmansperger2010robust; @Maronna2006robust].
@McKean1984comparison proposed a standard error estimator for the sample median from a continuous but not necessarily normal distribution, derived from a non-parametric confidence interval for the sample median. We use their approach to compute a MCSE for \(M_T\), the sample median of \(T\). Let \(c = \left\lceil(R + 1) / 2 - 1.96 \times \sqrt{R/4}\right\rfloor\), where the inner expression is rounded to the nearest integer. Then \[ MCSE\left(M_T\right) = \frac{T_{(R + 1 - c)} - T_{(c)}}{2 \times 1.96}. \tag{9.30} \] A Monte Carlo standard error for the median absolute deviation can be computed following the same approach, but substituting the order statistics of \(E_r = | T_r - \theta|\) in place of those for \(T_r\).
Trimmed mean bias with trimming proportion \(p\) is calculated by taking the mean of the the middle \((1 - 2p) \times R\) observations, which we have denoted as \(\tilde{T}_{\{p\}}\). A MCSE for the trimmed mean (and for the trimmed mean bias) is \[ MCSE\left(\tilde{T}_{\{p\}}\right) = \sqrt{\frac{U_p}{R}}, \tag{9.31} \] where \[ U_p = \frac{1}{(1 - 2p)R}\left( pR\left(T_{(pR)} - \tilde{T}_{\{p\}}\right)^2 + pR\left(T_{((1-p)R + 1)} - \tilde{T}_{\{p\}}\right)^2 + \sum_{r=pR + 1}^{(1 - p)R} \left(T_{(r)} - \tilde{T}_{\{p\}}\right)^2 \right) \] [@Maronna2006robust, Eq. 2.85].
Performance measures based on winsorization include winsorized bias, winsorized standard error, and winsorized RMSE. MCSEs for these measures can be computed using the same formuals as for the conventional measures of bias, empirical standard error, and RMSE, but using sample moments of \(\hat{X}_r\) in place of the sample moments of \(T_r\).
9.7.3 MCSE for Relative Variance Estimators
Estimating the MCSE of relative performance measures for variance estimators is complicated by the appearance of an estimated quantity in the denominator of the ratio. For instance, the relative bias of \(V\) is estimates as the ratio \(\bar{V} / S_T^2\), and both the numerator and denominator are estimated quantities that will include some Monte Carlo error. To properly account for the Monte Carlo uncertainty of the ratio, one possibility is to use formulas for the standard errors of ratio estimators. Alternately, we can use general uncertainty approximation techniques such as the jackknife or bootstrap [@boos2015Assessing]. The jackknife involves calculating a statistic of interest repeatedly, each time excluding one observation from the calculation. The variance of this set of one-left-out statistics then serves as a reasonable approximation to the actual sampling variance of the statistic calculated from the full sample.
To apply the jackknife to assess MCSEs of relative bias or relative RMSE of a variance estimator, we will need to compute several statistics repeatedly. Let \(\bar{V}_{(j)}\) and \(S_{T(j)}^2\) be the average variance estimate and the empirical variance estimate calculated from the set of replicates that excludes replicate \(j\), for \(j = 1,...,R\). The relative bias estimate, excluding replicate \(j\) would then be \(\bar{V}_{(j)} / S_{T(j)}^2\). Calculating all \(R\) versions of this relative bias estimate and taking the variance of these \(R\) versions yields a jackknife MCSE: \[ MCSE\left( \frac{ \bar{V}}{S_T^2} \right) = \sqrt{\frac{1}{R} \sum_{j=1}^R \left(\frac{\bar{V}_{(j)}}{S_{T(j)}^2} - \frac{\bar{V}}{S_T^2}\right)^2}. \tag{9.32} \] Similarly, a MCSE for the relative standard error of \(V\) is \[ MCSE\left( \frac{ S_V}{S_T^2} \right) = \sqrt{\frac{1}{R} \sum_{j=1}^R \left(\frac{S_{V(j)}}{S_{T(j)}^2} - \frac{S_V}{S_T^2}\right)^2}, \tag{9.33} \] where \(S_{V(j)}\) is the sample variance of \(V_1,...,V_R\), omitting replicate \(j\). To compute a MCSE for the relative RMSE of \(V\), we will need to compute the performance measure after omitting each observation in turn. Letting \[ RRMSE_{V} = \frac{1}{S_{T}^2}\sqrt{(\bar{V} - S_{T}^2)^2 + \frac{R - 1}{R} S_{V}^2} \] and \[ RRMSE_{V(j)} = \frac{1}{S_{T(j)}^2}\sqrt{(\bar{V}_{(j)} - S_{T(j)}^2)^2 + \frac{R - 1}{R} S_{V(j)}^2}, \] a jackknife MCSE for the estimated relative RMSE of \(V\) is \[ MCSE\left( RRMSE_{V} \right) = \sqrt{\frac{1}{R} \sum_{j=1}^R \left(RRMSE_{V(j)} - RRMSE_{V}\right)^2}. \tag{9.34} \]
Jackknife calculation would be cumbersome if we did it by brute force. However, a few algebra tricks provide a much quicker way. The tricks come from observing that \[ \begin{aligned} \bar{V}_{(j)} &= \frac{1}{R - 1}\left(R \bar{V} - V_j\right) \\ S_{V(j)}^2 &= \frac{1}{R - 2} \left[(R - 1) S_V^2 - \frac{R}{R - 1}\left(V_j - \bar{V}\right)^2\right] \\ S_{T(j)}^2 &= \frac{1}{R - 2} \left[(R - 1) S_T^2 - \frac{R}{R - 1}\left(T_j - \bar{T}\right)^2\right] \end{aligned} \] These formulas can be used to avoid re-computing the mean and sample variance from every subsample. Instead, all we need to do is calculate the overall mean and overall variance, and then do a small adjustment with each jackknife iteration.
Jackknife methods are useful for approximating MCSEs of other performance measures beyond just those for variance estimators. For instance, the jackknife is a convenient alternative for computing the MCSE of the empirical standard error or (raw) RMSE of a point estimator, which avoids the need to compute skewness or kurtosis. However, @boos2015Assessing notes that the jackknife does not work for performance measures involving medians, although bootstrapping remains valid.
9.7.4 MCSE for Confidence Intervals and Hypothesis Tests
Performance measures for confidence intervals and hypothesis tests are simple compared to those we have described for point and variance estimators. For evaluating hypothesis tests, the main measure is the rejection rate of the test, which is a proportion estimated as \(r_\alpha\) (Equation (9.20)). A MCSE for the estimated rejection rate is \[ MCSE(r_\alpha) = \sqrt{\frac{r_\alpha ( 1 - r_\alpha)}{R}}. \tag{9.35} \] This MCSE uses the estimated rejection rate to approximate its Monte Carlo error. When evaluating the validity of a test, we may expect the rejection rate to be fairly close to the nominal \(\alpha\) level, in which case we could compute a MCSE using \(\alpha\) in place of \(r_\alpha\), taking \(\sqrt{\alpha(1 - \alpha) / R}\). When evaluating power, we will not usually know the neighborhood of the rejection rate in advance of the simulation. However, a conservative upper bound on the MCSE can be derived by observing that MCSE is maximized when \(\rho_\alpha = \frac{1}{2}\), and so \[ MCSE(r_\alpha) \leq \sqrt{\frac{1}{4 R}}. \]
When evaluating confidence interval performance, we focus on coverage rates and expected widths. MCSEs for the estimated coverage rate work similarly to those for rejection rates. If the coverage rate is expected to be in the neighborhood of the intended coverage level \(\beta\), then we can approximate the MCSE as \[ MCSE(\widehat{\text{Coverage}}(A,B)) = \sqrt{\frac{\beta(1 - \beta)}{R}}. \tag{9.36} \] Alternately, Equation (9.36) could be computed using the estimated coverage rate \(\widehat{\text{Coverage}}(A,B)\) in place of \(\beta\).
Finally, the expected confidence interval width can be estimated as \(\bar{W}\), with MCSE \[ MCSE(\bar{W}) = \sqrt{\frac{S_W^2}{R}}, \tag{9.37} \] where \(S_W^2\) is the sample variance of \(W_1,...,W_R\), the widths of the confidence interval from each replication.
9.7.5 Calculating MCSEs With the simhelpers Package
The simhelpers package provides several functions for calculating most of the performance measures that we have reviewed, along with MCSEs for each performance measures.
The functions are easy to use.
Consider this set of simulation runs on the Welch dataset:
library( simhelpers )
data( welch_res )
welch <-
welch_res %>%
dplyr::select(-seed, -iterations ) %>%
mutate(method = case_match(method, "Welch t-test" ~ "Welch", .default = method))
head(welch)## # A tibble: 6 × 9
## n1 n2 mean_diff method est var
## <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
## 1 50 50 0 t-test 0.0258 0.0954
## 2 50 50 0 Welch 0.0258 0.0954
## 3 50 50 0 t-test 0.00516 0.0848
## 4 50 50 0 Welch 0.00516 0.0848
## 5 50 50 0 t-test -0.0798 0.0818
## 6 50 50 0 Welch -0.0798 0.0818
## # ℹ 3 more variables: p_val <dbl>,
## # lower_bound <dbl>, upper_bound <dbl>We can calculate performance measures across all the range of scenarios. Here is the rejection rate for the traditional \(t\)-test based on the subset of simulation results with sample sizes of \(n_1 = n_2 = 50\) and a mean difference of 0, using \(\alpha\) levels of .01 and .05:
welch_sub <- filter(welch, method == "t-test", n1 == 50, n2 == 50, mean_diff == 0 )
calc_rejection(welch_sub, p_values = p_val, alpha = c(.01, .05))## K_rejection rej_rate_01 rej_rate_05
## 1 1000 0.009 0.048
## rej_rate_mcse_01 rej_rate_mcse_05
## 1 0.002986469 0.006759882The column labeled K_rejection reports the number of replications used to calculate the performance measures.
Here is the coverage rate calculated for the same condition:
calc_coverage(
welch_sub,
lower_bound = lower_bound, upper_bound = upper_bound,
true_param = mean_diff
)## # A tibble: 1 × 5
## K_coverage coverage coverage_mcse width
## <int> <dbl> <dbl> <dbl>
## 1 1000 0.952 0.00676 1.25
## # ℹ 1 more variable: width_mcse <dbl>The performance functions are designed to be used within a tidyverse-style workflow, including on grouped datasets. For instance, we can calculate rejection rates for every distinct scenario examined in the simulation:
all_rejection_rates <-
welch %>%
group_by( n1, n2, mean_diff, method ) %>%
summarise(
calc_rejection( p_values = p_val, alpha = c(.01, .05) )
)The resulting summaries are reported in table 9.1.
| n1 | n2 | mean_diff | method | K_rejection | rej_rate_01 | rej_rate_05 | rej_rate_mcse_01 | rej_rate_mcse_05 |
|---|---|---|---|---|---|---|---|---|
| 50 | 50 | 0.0 | Welch | 1000 | 0.009 | 0.047 | 0.003 | 0.007 |
| 50 | 50 | 0.0 | t-test | 1000 | 0.009 | 0.048 | 0.003 | 0.007 |
| 50 | 50 | 0.5 | Welch | 1000 | 0.157 | 0.335 | 0.012 | 0.015 |
| 50 | 50 | 0.5 | t-test | 1000 | 0.162 | 0.340 | 0.012 | 0.015 |
| 50 | 50 | 1.0 | Welch | 1000 | 0.677 | 0.871 | 0.015 | 0.011 |
| 50 | 50 | 1.0 | t-test | 1000 | 0.686 | 0.876 | 0.015 | 0.010 |
| 50 | 50 | 2.0 | Welch | 1000 | 1.000 | 1.000 | 0.000 | 0.000 |
| 50 | 50 | 2.0 | t-test | 1000 | 1.000 | 1.000 | 0.000 | 0.000 |
| 50 | 70 | 0.0 | Welch | 1000 | 0.008 | 0.039 | 0.003 | 0.006 |
| 50 | 70 | 0.0 | t-test | 1000 | 0.004 | 0.027 | 0.002 | 0.005 |
| 50 | 70 | 0.5 | Welch | 1000 | 0.202 | 0.426 | 0.013 | 0.016 |
| 50 | 70 | 0.5 | t-test | 1000 | 0.139 | 0.341 | 0.011 | 0.015 |
| 50 | 70 | 1.0 | Welch | 1000 | 0.820 | 0.937 | 0.012 | 0.008 |
| 50 | 70 | 1.0 | t-test | 1000 | 0.743 | 0.904 | 0.014 | 0.009 |
| 50 | 70 | 2.0 | Welch | 1000 | 1.000 | 1.000 | 0.000 | 0.000 |
| 50 | 70 | 2.0 | t-test | 1000 | 1.000 | 1.000 | 0.000 | 0.000 |
9.7.6 MCSE Calculation in our Cluster RCT Example
In Section 9.1.1, we computed performance measures for three point estimators of the school-level average treatment effect in a cluster RCT.
We can carry out the same calculations using the calc_absolute() function from simhelpers, which also provides MCSEs for each measure.
Examining the MCSEs is useful to ensure that 1000 replications of the simulation is suffiicent to provide reasonably precise estimates of the performance measures.
In particular, we have:
library( simhelpers )
runs %>%
group_by(method) %>%
summarise(
calc_absolute(
estimates = ATE_hat, true_param = ATE,
criteria = c("bias","stddev", "rmse")
)
)## # A tibble: 3 × 8
## method K_absolute bias bias_mcse stddev
## <chr> <int> <dbl> <dbl> <dbl>
## 1 Agg 1000 -0.000166 0.00676 0.214
## 2 LR 1000 0.0824 0.00707 0.224
## 3 MLM 1000 0.0122 0.00674 0.213
## # ℹ 3 more variables: stddev_mcse <dbl>,
## # rmse <dbl>, rmse_mcse <dbl>We see the MCSEs are quite small relative to the linear regression bias term and all the SEs (stddev) and RMSEs. Results based on 1000 replications seems adequate to support our conclusions about the gross trends identified.
We have not simulated enough to rule out the possibility that the aggregation estimator and multilevel modeling estimator could be slightly biased. Given our MCSEs, they could have true bias of as much as 0.01 (two MCSEs).
9.8 Summary of Peformance Measures
We list most of the performance criteria we saw in this chapter in the table below, for reference:
| Criterion | Definition | Estimator | Monte Carlo Standard Error |
|---|---|---|---|
| Measures for point estimators | |||
| Bias | \(\E(T) - \theta\) | \(\bar{T} - \theta\) | (9.24) |
| Median bias | \(\M(T) - \theta\) | \(m_T - \theta\) | (9.30) |
| Trimmed bias | |||
| Variance | \(\E\left[\left(T - \text{E}(T)\right)^2\right]\) | \(S_T^2\) | (9.25) |
| Standard error | \(\sqrt{\E\left[\left(T - \text{E}(T)\right)^2\right]}\) | \(S_T\) | (9.26) |
| Mean squared error | \(\E\left[\left(T - \theta\right)^2\right]\) | \(\left(\bar{T} - \theta\right)^2 + \frac{R - 1}{R}S_T^2\) | (9.27) |
| Root mean squared error | \(\sqrt{\E\left[\left(T - \theta\right)^2\right]}\) | \(\sqrt{\left(\bar{T} - \theta\right)^2 + \frac{R - 1}{R} S_T^2}\) | (9.28) |
| Median absolute error | \(\M\left[\left|T - \theta\right|\right]\) | \(\left[\left|T - \theta\right|\right]_{R/2}\) | (9.30) |
| Relative bias | \(\E(T) / \theta\) | \(\bar{T} / \theta\) | (9.29) |
| Relative median bias | \(\M(T) / \theta\) | \(m_T / \theta\) | (9.29) |
| Relative RMSE | \(\sqrt{\E\left[\left(T - \theta\right)^2\right]} / \theta\) | \(\frac{\sqrt{\left(\bar{T} - \theta\right)^2 + \frac{R - 1}{R}S_T^2}}{\theta}\) | (9.29) |
- Bias and median bias are measures of whether the estimator is systematically higher or lower than the target parameter.
- Variance is a measure of the precision of the estimator—that is, how far it deviates from its average. We might look at the square root of this, to assess the precision in the units of the original measure. This is the true SE of the estimator.
- Mean-squared error is a measure of overall accuracy, i.e. is a measure how far we typically are from the truth. We more frequently use the root mean squared error, or RMSE, which is just the square root of the MSE.
- The median absolute deviation (MAD) is another measure of overall accuracy that is less sensitive to outlier estimates. The RMSE can be driven up by a single bad egg. The MAD is less sensitive to this.
9.9 Concluding thoughts
In practice, many data analysis procedures produce multiple pieces of information—not just point estimates, but also standard errors and confidence intervals and p-values from null hypothesis tests—and those pieces are inter-related.
For instance, a confidence interval is usually computed from a point estimate and its standard error.
Consequently, the performance of that confidence interval will be strongly affected by whether the point estimator is biased and whether the standard error tends to understates or over-states the true uncertainty.
Likewise, the performance of a hypothesis testing procedure will often strongly depend on the properties of the point estimator and standard error used to compute the test.
Thus, most simulations will involve evaluating a data analysis procedure on several measures to arrive at a holistic understanding of its performance.
Moreover, the main aim of many simulations is to compare the performance of several different estimators or to determine which of several data analysis procedures is preferable. For such aims, we will need to use the performance measures to understand whether a set of procedures work differently, when and how one is superior to the other, and what factors influence differences in performance. To fully understand the advantages and trade-offs among a set of estimators, we will generally need to compare them using several performance measures.
9.10 Exercises
9.10.1 Brown and Forsythe (1974) results
Use the
generate_ANOVA_datadata-generating function for one-way heteroskedastic ANOVA (Section 5.1) and the data-analysis function you wrote for Exercise 7.5.1 to create a simulation driver function for the Brown and Forsythe simulations.Use your simulation driver to evaluate the Type-I error rate of the ANOVA \(F\)-test, Welch’s test, and the BFF* test for a scenario with four groups, sample sizes \(n_1 = 11\), \(n_2 = 16\), \(n_3 = 16\), \(n_4 = 21\), equal group means \(\mu_1 = \mu_2 = \mu_3 = \mu_4 = 0\), and group standard deviations \(\sigma_1 = 3\), \(\sigma_2 = 2\), \(\sigma_3 = 2\), \(\sigma_4 = 1\). Are all of the tests level-\(\alpha\)?
Use your simulation driver to evaluate the power of each test for a scenario with group means of \(\mu_1 = 0\), \(\mu_2 = 0.2\), \(\mu_3 = 0.4\), \(\mu_4 = 0.6\), with sample sizes and group standard deviations as listed above. Which test has the highest power?
9.10.2 Size-adjusted power
When different hypothesis testing procedures have different rejection rates under the null hypothesis, it becomes difficult to interpret differences in the non-null power of the tests. One approach for conducting a fair comparison of testing procedures in this situation is to compute the size-adjusted power of the tests. Size-adjusted power involves computing the rejection rate of a test using a different threshold \(\alpha'\), selected so that the Type-I error rate of the test is equal to the desired \(\alpha\) level. Specifically, size adjusted power is \[ \rho^{adjusted}_\alpha(\theta) = \Prob(P < \rho_\alpha(0)). \] To estimate size-adjusted power using simulation, we first need to estimate the Type-I error rate, \(r_\alpha(0)\). We can then evaluate the rejection rate of the testing procedure under scenarios with other values of \(\theta\) by computing \[ r^{adjusted}_\alpha(\theta) = \frac{1}{R} \sum_{r=1}^R I(P_r < r_{\alpha}(0)). \]
Compute the size-adjusted power of the ANOVA \(F\)-test, Welch’s test, and the BFF* test for the scenario in part (3) of Exercise 9.10.1.
9.10.3 Three correlation estimators
Consider the bivariate negative binomial distribution model described in Exercise 6.9.6. Suppose that we want to estimate the correlation parameter \(\rho\) of the latent bivariate normal distribution. Without studying the statistical theory for this problem, we might think to use simulation to evaluate whether any common correlation measures work well for estimating this parameter. Potential candidate estimators include the usual sample Pearson’s correlation, Spearman’s rank correlation, or Kendall’s \(\tau\) coefficient. The latter two estimators might seem promising because they are based on the ranked data, so could be more appropriate that Pearson’s correlation for frequency count variates.
The following estimation function computes all three correlations, along with corresponding \(p\)-values for the null hypothesis of no association and confidence intervals computed using Fisher’s \(z\) transformation. The confidence interval calculations are developed for Pearson’s correlation under bivariate normality, so they might not be appropriate for this data-generating process or for Spearman’s or Kendall’s correlations. Still, we can use simulation to see how well or how poorly they perform.
three_corrs <- function(
data,
method = c("pearson","kendall","spearman"),
level = 0.95
) {
r_est <- lapply(method, \(m) cor.test(data$C1, data$C2, method = m, exact = FALSE))
est <- sapply(r_est, \(x) as.numeric(x$estimate))
pval <- sapply(r_est, \(x) x$p.value)
z_est <- atanh(est)
se_z <- 1 / sqrt(nrow(data) - 3)
crit <- qnorm(1 - (1 - level) / 2)
ci_lo <- tanh(z_est - crit * se_z)
ci_hi <- tanh(z_est + crit * se_z)
data.frame(
stat = method,
r = est,
z = z_est,
se_z = se_z,
pval = pval,
ci_lo = ci_lo,
ci_hi = ci_hi
)
}Combine your data-generating function and
three_corrs()into a simulation driver.Use your simulation driver to generate 500 replications of the simulation for a scenario with \(N = 20\), \(\mu_1 = \mu_2 = 5\), \(p_1 = p_2 = 0.5\), and \(\rho = 0.7\).
Compute the bias, empirical standard error, and RMSE of each correlation estimator, along with corresponding MCSEs. Which correlation estimator is most accurate?
Compute the coverage rate and expected width of the confidence intervals based on each correlation estimator. Do any of the estimators have reasonable coverage? If so, which has the best expected width?
Use your simulation driver to estimate the Type-I error rate of the hypothesis tests for each correlation coefficient for a scenario with \(N = 20\), \(\mu_1 = \mu_2 = 5\), \(p_1 = p_2 = 0.5\). Are any of the tests level-\(\alpha\)?
9.10.4 Confidence interval comparison
Consider the estimation functions for the cluster RCT example, as given in Section 7.2.
Modify the functions to return both normal Wald-type 95% confidence intervals (as computed in Section 9.3.1) and cluster-robust confidence intervals based on \(t\) distributions with Satterthwaite degrees of freedom.
For the latter, use conf_int() from the clubSandwich package, as in the following example code:
library(clubSandwich)
M1 <- lme4::lmer(
Yobs ~ 1 + Z + (1 | sid),
data = dat
)
conf_int(M1, vcov = "CR2")
M2 <- estimatr::lm_robust(
Yobs ~ 1 + Z, data = dat,
clusters = sid, se_type = se_type
)
conf_int(M2, cluster = dat$sid, vcov = "CR2")
M3 <- estimatr::lm_robust(
Ybar ~ 1 + Z, data = datagg,
se_type = se_type
)
conf_int(M3, cluster = dat$sid, vcov = "CR2")Pick some simulation parameters and estimate the coverage and interval width of both types of confidence intervals. How do the normal Wald-type intervals compare to the cluster-robust intervals?
9.10.5 Jackknife calculation of MCSEs for RMSE
The following code generates 100 replications of a simulation of three average treatment effect estimators in a cluster RCT, using a simulation driver function we developed in Section 8.3 using components described in Sections 6.6 and 7.2.
set.seed( 20251029 )
runs_val <- sim_cluster_RCT(
reps = 100,
J = 16, n_bar = 20, alpha = 0.5,
gamma_1 = 0.3, gamma_2 = 0.8,
sigma2_u = 0.25, sigma2_e = 0.75
)Compute the RMSE of each estimator, and use the jackknife technique described in Section 9.7.3 to compute a MCSE for the RMSE.
Check your results against the results from calc_absolute() in the simhelpers package.
9.10.6 Jackknife calculation of MCSEs for RMSE ratios
Continuing from Exercise 9.10.5, compute the ratio of the RMSE of each estimator to the RMSE of the linear regression estimator. Use the jackknife technique to compute a MCSE for these RMSE ratios.
9.10.7 Distribution theory for person-level average treatment effects
Section 6.6 described a data-generating process for a cluster-randomized experiment in which the school-specific treatment effects varied according to the size of the school. The auxiliary model for the size of school \(j\) was \[ n_j \sim \text{Unif}\left[ (1-\alpha)\bar{n}, (1+\alpha)\bar{n} \right], \] where \(\bar{n}\) was the average school size and \(0 \leq \alpha < 1\) determines the degree of variation in school sizes. The data-generating process for the outcome data was \[ Y_{ij} = \gamma_{0} + \gamma_{1} Z_j + \gamma_{2} Z_j S_j + u_j + \epsilon_{ij}, \] where \(Y_{ij}\) is the outcome for student \(i\) in school \(j\), \(Z_j\) is an indicator for whether school \(j\) is assigned to treatment \((Z_j = 1)\) or control \((Z_j = 0)\), and \(S_j = \frac{n_j - \bar{n}}{\bar{n}}\). The error terms \(u_j\) and \(e_{ij}\) are both assumed to be normally distributed with zero means.
Under this model, the average treatment effect for school \(j\) is \(\tau_j = \gamma_1 + \gamma_2 S_j\). Because \(\E(S_j) = 0\) by construction, the average of the school-specific treatment effects is \(\gamma_1\). This is the school-level population average treatment effect estimate. But what is the student-level population average treatment effect estimate? Use the properties of the uniform distribution to find the student-level population average treatment effect \(\E\left( \frac{n_j}{\bar{n}} \times \tau_j \right)\). Check your derivation by simulating a large sample of school sizes and school-specific treatment effects.
Generally, when people say “Standard Error” they actually mean estimated Standard Error, (\(\widehat{SE}\)), as we would calculate in a real data analysis (where we have only a single realization of the data-generating process). It is easy to forget that this standard error is itself an estimate of a parameter–the true or empirical SE—and thus has its own uncertainty.↩︎
For a normally distributed sampling distribution, the interquartile range is 1.35 SD; with \(w = 2\), the lower and upper thresholds would then fall at \(\pm 3.37\) SD, or the \(0.04^{th}\) and \(99.96^{th}\) percentiles. Still assuming a normal sampling distribution, taking \(w = 2.5\) will mean that the thresholds fall at the \(0.003^{th}\) and \(99.997^{th}\) percentiles.↩︎
See the delightfully titled section 11.5, “The Joke Is on Us: The Standard Deviation Estimator is Biased after All,” in @westfall2013understanding for further discussion.↩︎
The convention of using \(\alpha = .05\) does not have a strong theoretical rationale. Many scholars have criticized the rote application of this convention and argued for using other \(\alpha\) levels. See @Benjamin2017redefine and @Lakens2018justify for spirited arguments about choosing \(\alpha\) levels for hypothesis testing.↩︎
In the cluster-RCT example, the distribution theory is tractable. See Exercise 9.10.7↩︎
To be precise, the formulas that we give are estimators for the Monte Carlo standard errors of the performance measure estimators. Our presentation does not emphasize this point because the performance measures will usually be estimated using a large number of replications from an independent and identically distributed process, so the distinction between empirical and estimated standard errors will not be consequential.↩︎
The delta method approximation says (with some conditions), that if we assume \(X \sim N\left( \phi, \sigma_X^2 \right)\), then we can approximate the distribution of \(g(X)\) for some continuous function \(g(\cdot)\) as \[ g(X) \sim N\left( g(\phi), \;\; g'(\phi)^2 \times \sigma_X^2 \right),\] where \(g'(\phi)\) is the derivative of \(g(\cdot)\) evaluated at \(\phi\). Following this approximation, \[ SE( g(X) ) \approx \left| g'(\theta) \right| \times SE(X) .\] For estimation, we plug in \(\hat{\theta}\) and our estimate of \(SE(X)\) into the above. To find the MCSE for \(S_T\), we can apply the delta method approximation to \(X = S_T^2\) with \(g(x) = \sqrt(x)\) and \(g'(x) =\frac{1}{2\sqrt{x}}\).↩︎